m140002 (PDF)
File information
This PDF 1.4 document has been generated by / PDFill: Free PDF Writer and Tools, and has been sent on pdf-archive.com on 27/07/2015 at 13:56, from IP address 103.58.x.x.
The current document download page has been viewed 616 times.
File size: 270.04 KB (9 pages).
Privacy: public file



File preview
Biojournal of Science and Technology
Research Article
In silico miRNA Target Identification within the Human
Peroxisome Proliferator -Activated
Activated Receptor Gamma (PPARG)
Gene
Sudip Paul*, Moumoni Saha, Kazi Saiful Islam, Md. Yeashin Gazi, Sohel Ahmed
Jahangirnagar University, Savar, Dhaka 1342, Bangladesh
*Corresponding author
Sudip Paul, Dept. of Biochemistry and Molecular Biology,
Jahangirnagar University, Savar, Dhaka 1342,
Bangladesh. Mob: 01674389745.
e-mail: sudippaul.bcmb@gmail.com
Published: 25-09-2014
Biojournal of Science and Technology Vol.1:2014
Academic Editor: Dr. Mohammad Nazmul Ahsan
Received: 17-07-2014
2014
Accepted: 13-08-2014
2014
Article no: m140002
This is an Open Access article distributed under the terms of the Creative Commons Attribution License
(http://creativecommons.org/licenses/by/4.0
http://creativecommons.org/licenses/by/4.0 ), which permits unrestricted use, distribution, and
reproduction in any medium, provided the original work is properly cited.
Abstract
MicroRNAs (miRNAs),
), an abundant class of 21
21-25 nucleotides long non-coding
coding RNAs, regulate
eukaryotic gene expression and therefore implicated in a wide range of biological processes. The miRNAmiRNA
related genetic alterations are possibly more implicated in human diseases than currently appreciated.
miRNA target prediction using bioinformatics tools is often the first line approach in studying gene
regulation. Such predictions will help in setting search priorities for experimental validation of gene
controlling mechanisms. But finding a functional miRNA target in the human genome yet remains a
challenging task. In the present study, miRNA target sites within the complete sequences (5′
(5 UTR, CDS
and 3′ UTR) of human PPARG gene were investigated using miRwalk database. We found 26,
26 52 and 85
different miRNA target sites within the 55′ UTR, CDS and 3′ UTR regions of the gene, respectively. This
computational approach will subsequently allow better in vitro confirmation of the miRNA regulatory
networks in cellular systems.
Keywords: microRNA, In silico
silico, target site, PPARG, miRWalk
ISSN 2410-9754
Vol:1, 2014
INTRODUCTION
(Srinivasan et al 2013). Therefore, microRNAs
MicroRNAs (miRNAs) are a broad class of
displaying deregulated expression in the context of
naturally occurring small non-coding RNAs of
specific diseases are of particular interest as
about 21-25 nucleotides in length and found in
therapeutic targets especially if they can be shown
plants, animals and some viruses. The main
to coordinate such disease networks.
functions of miRNAs are to down-regulate gene
expression in translational repression, cleavage of
Peroxisome
proliferators-activated
messenger RNA (mRNA) and in a variety of other
gamma (PPARγ or PPARG) encoded by the
biological processes. Each miRNA is partially or
PPARG gene in humans belongs to the nuclear
completely complementary to one or more mRNAs
hormone receptor superfamily of ligand-activated
(Friedman et al. 2009, Landgraf et al. 2007).
transcription factors and originally has been
Transcription of miRNAs occurs through RNA
characterized to be important for adipogenesis and
polymerase II9 and subsequent processing is
glucose metabolism. There are two isoforms
mediated by the nuclear ribonuclease III (RNase
described (PPARG 1 and -2) (Vidal-Puig A. J. et al.
III) enzyme Drosha to form precursor miRNAs
1997). PPARG has been associated with various
(70–100 nucleotides). Following transportation to
diseases including obesity, diabetes mellitus,
the cytoplasm by exportin 5, a further cleavage
atherosclerosis, and cancer. PPARG agonists have
occurs via another RNase III enzyme, Dicer, to
been
form the mature miRNA (He and Hannon 2004,
of hyperlipidaemia and hyperglycemia (Li et al.
Zeng and Cullen 2006).
2008).
used
PPARG
in
is
the
important
receptor
treatment
to
shape
an
anti-inflammatory macrophage phenotype and
miRNAs
modulate
both
physiological
and
appears crucial for dampening inflammation
pathological pathways by post-transcriptionally
(Rosen et al. 1999). miRNAs have been reported to
inhibiting the expression of a plethora of target
destabilize PPARG mRNA which can lead to
genes. miRNAs deregulate gene expression mostly
impaired PPARG abundance (Schoonjans et al.
by imperfect binding to complementary sites
1996, Vidal-Puig A. et al. 1996). Therefore,
within transcript sequences and suppresses their
miRNA target site identification within the PPARG
translation, stimulate their de-adenylation and
gene is quite important in studying PPARG gene
degradation or induce their cleavage (Bartel 2004,
regulation.
Perron and Provost 2008).
There are a number of miRNA target prediction
The decisive regulatory functions exhibited by the
algorithms exploiting different approaches have
miRNA are found to be associated with a wide
been recently developed, and many methods of
variety of human diseases such as cancer, heart
experimental validation have been premeditated.
diseases, metabolic disorders, neurodegenerative
However, it is difficult to predict miRNA targets
disorders etc. as reviewed by Srinivasan et al.
within the animal genomes due to its partial
@2014, GNP
Biojournal of Science and Technology
Pa g e |1
ISSN 2410-9754
Vol:1, 2014
complementation to their target mRNA (Martin et
several web-based or non web-based computer
al. 2007). For this shortcoming, the interactions of
software programs for predicting miRNAs and
miRNA with their mRNA counterparts are
their targets have been developed in order to
complex and poorly understood. In the study in
predict
silico based miRNA targets identification within
validation. Even though many computational
the human PPARG gene was performed.
methods for the identification of miRNA may have
targets
for
follow
up
experimental
their own limitations, there is no other option now
METHODS
other than to use computational methods for
The miRWalk, a comprehensive database of
miRNA predictions. The next step in miRNA
miRNA from human, mouse and rat was used to
research is to identify and experimentally validate
identify miRNA target sites within the human
their mRNA targets.
PPARG gene based on a comparison of identified
All computer-based miRNA target prediction
miRNA binding sites with the 8 established
programs are based on specific parameters where
miRNA-target prediction programs i.e. RNA22,
slight variation results for the same target input.
miRanda, miRDB, TargetScan, RNA- hybrid,
Such weakness of single in silico studies can be
PITA, PICTAR, and Diana-microT (Dweep et al.
partially compensated by predicting targets using
2011). The miRWalk algorithm identifies the
multiple
programs.
longest
dynamic
programming
consecutive
complementary
between
Scoring
(John
methods
et
al.
using
2004,
miRNA and gene sequences. miRWalk was used
Kiriakidou et al. 2004, Lewis et al. 2003) and a
for investigating predicted targets of microRNAs
complementarily-based strategy (Lewis et al. 2003,
in the complete sequences (5′ UTR, CDS and 3′
Rajewsky and Socci 2004) are generally preferred
UTR) of PPARG gene in the human genome.
to rank the prediction results. These approaches
Default parameters were used regarding minimum
have been quite successful for a few top ranked
seed length (7) and p value (0.05).
results. miRNAs targets calculated from multiple
prediction methods significantly improved target
RESULTS AND DISCUSSION
prediction accuracy. Therefore, 8 key programs
Because of the several limitations associated with
were used in the present study to optimize our
genetic screening and experimental approaches for
search and to unravel miRNA target sequences of
discovering founding members of miRNAs such as
the PPARG gene cluster with high accuracy.
low efficiency, time consuming and high cost,
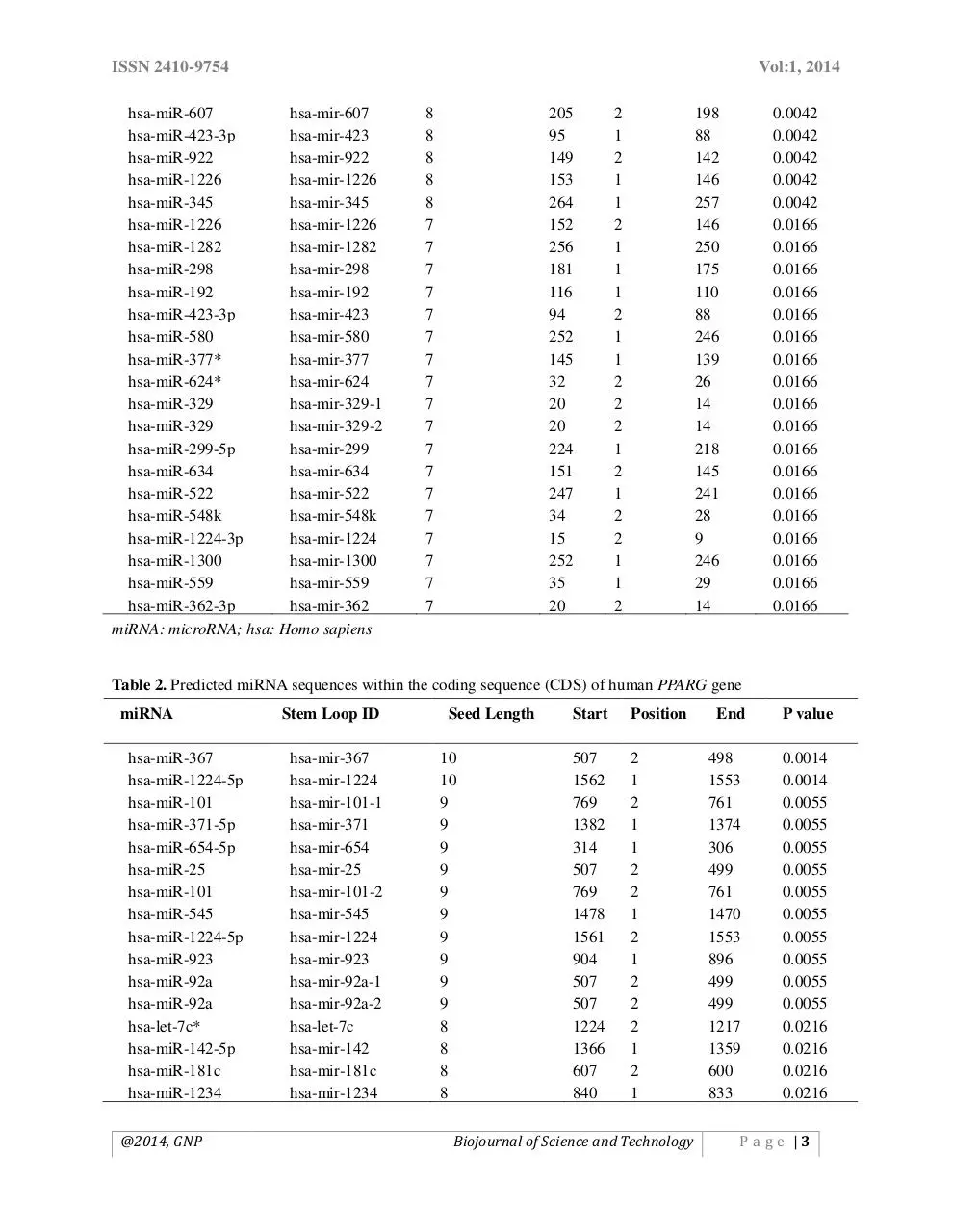
Table 1. Predicted miRNA sequences within the 5′-untranslated region (5′-UTR) of human PPARG gene
miRNA
hsa-miR-181a-2*
hsa-miR-345
hsa-miR-181a-2*
@2014, GNP
Stem Loop ID
hsa-mir-181a-2
hsa-mir-345
hsa-mir-181a-2
Seed Length
Start
Position
End
10
9
9
120
75
119
1
2
2
111
67
111
Biojournal of Science and Technology
P value
0.0003
0.0010
0.0010
Pa g e |2
ISSN 2410-9754
Vol:1, 2014
hsa-miR-607
hsa-mir-607
hsa-miR-423-3p
hsa-mir-423
hsa-miR-922
hsa-mir-922
hsa-miR-1226
hsa-mir-1226
hsa-miR-345
hsa-mir-345
hsa-miR-1226
hsa-mir-1226
hsa-miR-1282
hsa-mir-1282
hsa-miR-298
hsa-mir-298
hsa-miR-192
hsa-mir-192
hsa-miR-423-3p
hsa-mir-423
hsa-miR-580
hsa-mir-580
hsa-miR-377*
hsa-mir-377
hsa-miR-624*
hsa-mir-624
hsa-miR-329
hsa-mir-329-1
hsa-miR-329
hsa-mir-329-2
hsa-miR-299-5p
hsa-mir-299
hsa-miR-634
hsa-mir-634
hsa-miR-522
hsa-mir-522
hsa-miR-548k
hsa-mir-548k
hsa-miR-1224-3p
hsa-mir-1224
hsa-miR-1300
hsa-mir-1300
hsa-miR-559
hsa-mir-559
hsa-miR-362-3p
hsa-mir-362
miRNA: microRNA; hsa: Homo sapiens
8
8
8
8
8
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
205
95
149
153
264
152
256
181
116
94
252
145
32
20
20
224
151
247
34
15
252
35
20
2
1
2
1
1
2
1
1
1
2
1
1
2
2
2
1
2
1
2
2
1
1
2
198
88
142
146
257
146
250
175
110
88
246
139
26
14
14
218
145
241
28
9
246
29
14
0.0042
0.0042
0.0042
0.0042
0.0042
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
0.0166
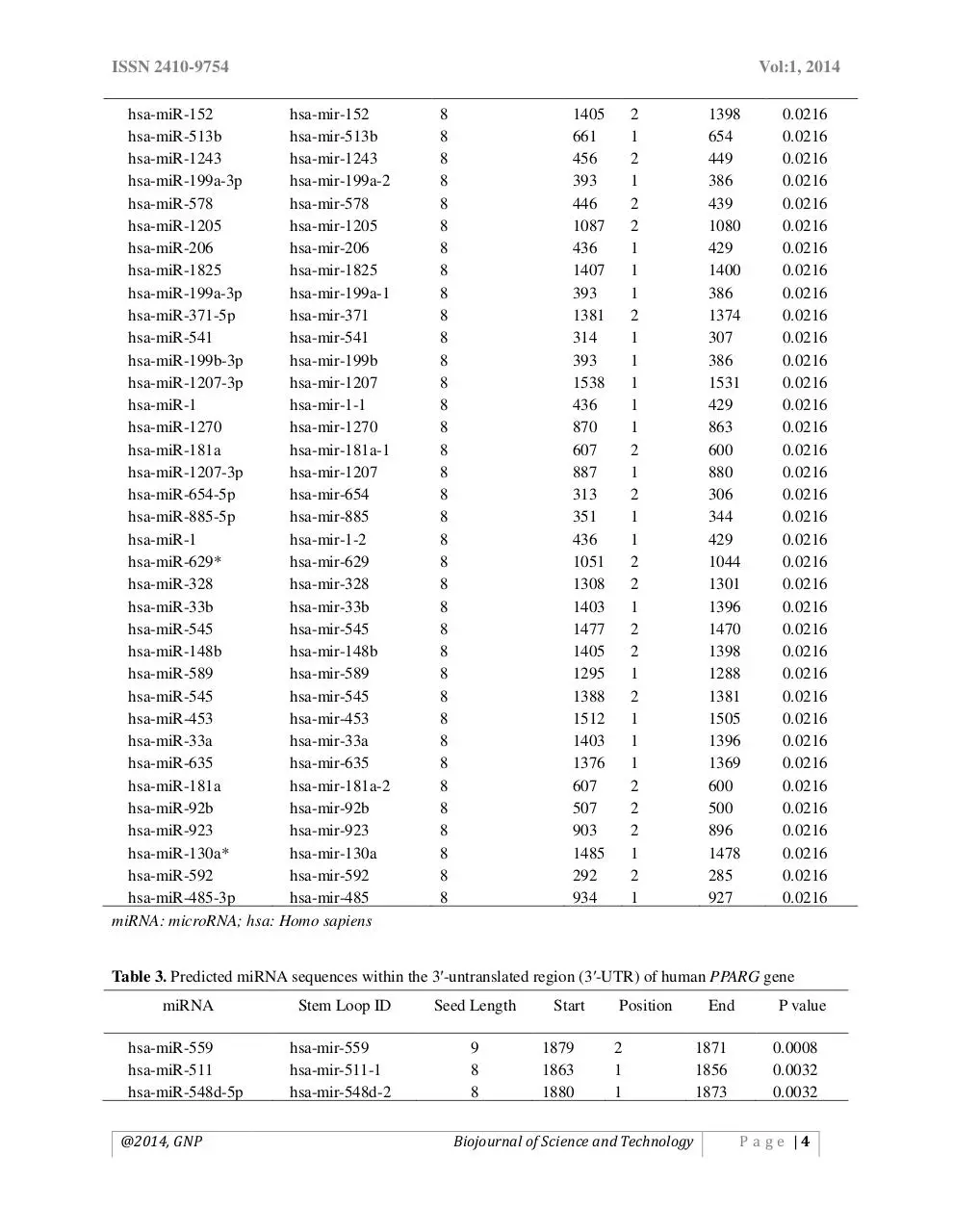
Table 2. Predicted miRNA sequences within the coding sequence (CDS) of human PPARG gene
miRNA
hsa-miR-367
hsa-miR-1224-5p
hsa-miR-101
hsa-miR-371-5p
hsa-miR-654-5p
hsa-miR-25
hsa-miR-101
hsa-miR-545
hsa-miR-1224-5p
hsa-miR-923
hsa-miR-92a
hsa-miR-92a
hsa-let-7c*
hsa-miR-142-5p
hsa-miR-181c
hsa-miR-1234
@2014, GNP
Stem Loop ID
hsa-mir-367
hsa-mir-1224
hsa-mir-101-1
hsa-mir-371
hsa-mir-654
hsa-mir-25
hsa-mir-101-2
hsa-mir-545
hsa-mir-1224
hsa-mir-923
hsa-mir-92a-1
hsa-mir-92a-2
hsa-let-7c
hsa-mir-142
hsa-mir-181c
hsa-mir-1234
Seed Length
10
10
9
9
9
9
9
9
9
9
9
9
8
8
8
8
Start
Position
507
1562
769
1382
314
507
769
1478
1561
904
507
507
1224
1366
607
840
2
1
2
1
1
2
2
1
2
1
2
2
2
1
2
1
Biojournal of Science and Technology
End
498
1553
761
1374
306
499
761
1470
1553
896
499
499
1217
1359
600
833
P value
0.0014
0.0014
0.0055
0.0055
0.0055
0.0055
0.0055
0.0055
0.0055
0.0055
0.0055
0.0055
0.0216
0.0216
0.0216
0.0216
Pa g e |3
ISSN 2410-9754
Vol:1, 2014
hsa-miR-152
hsa-mir-152
hsa-miR-513b
hsa-mir-513b
hsa-miR-1243
hsa-mir-1243
hsa-miR-199a-3p
hsa-mir-199a-2
hsa-miR-578
hsa-mir-578
hsa-miR-1205
hsa-mir-1205
hsa-miR-206
hsa-mir-206
hsa-miR-1825
hsa-mir-1825
hsa-miR-199a-3p
hsa-mir-199a-1
hsa-miR-371-5p
hsa-mir-371
hsa-miR-541
hsa-mir-541
hsa-miR-199b-3p
hsa-mir-199b
hsa-miR-1207-3p
hsa-mir-1207
hsa-miR-1
hsa-mir-1-1
hsa-miR-1270
hsa-mir-1270
hsa-miR-181a
hsa-mir-181a-1
hsa-miR-1207-3p
hsa-mir-1207
hsa-miR-654-5p
hsa-mir-654
hsa-miR-885-5p
hsa-mir-885
hsa-miR-1
hsa-mir-1-2
hsa-miR-629*
hsa-mir-629
hsa-miR-328
hsa-mir-328
hsa-miR-33b
hsa-mir-33b
hsa-miR-545
hsa-mir-545
hsa-miR-148b
hsa-mir-148b
hsa-miR-589
hsa-mir-589
hsa-miR-545
hsa-mir-545
hsa-miR-453
hsa-mir-453
hsa-miR-33a
hsa-mir-33a
hsa-miR-635
hsa-mir-635
hsa-miR-181a
hsa-mir-181a-2
hsa-miR-92b
hsa-mir-92b
hsa-miR-923
hsa-mir-923
hsa-miR-130a*
hsa-mir-130a
hsa-miR-592
hsa-mir-592
hsa-miR-485-3p
hsa-mir-485
miRNA: microRNA; hsa: Homo sapiens
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
1405
661
456
393
446
1087
436
1407
393
1381
314
393
1538
436
870
607
887
313
351
436
1051
1308
1403
1477
1405
1295
1388
1512
1403
1376
607
507
903
1485
292
934
2
1
2
1
2
2
1
1
1
2
1
1
1
1
1
2
1
2
1
1
2
2
1
2
2
1
2
1
1
1
2
2
2
1
2
1
1398
654
449
386
439
1080
429
1400
386
1374
307
386
1531
429
863
600
880
306
344
429
1044
1301
1396
1470
1398
1288
1381
1505
1396
1369
600
500
896
1478
285
927
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
0.0216
Table 3. Predicted miRNA sequences within the 3′-untranslated region (3′-UTR) of human PPARG gene
miRNA
Stem Loop ID
Seed Length
hsa-miR-559
hsa-miR-511
hsa-miR-548d-5p
hsa-mir-559
hsa-mir-511-1
hsa-mir-548d-2
9
8
8
@2014, GNP
Start
1879
1863
1880
Position
2
1
1
Biojournal of Science and Technology
End
1871
1856
1873
P value
0.0008
0.0032
0.0032
Pa g e |4
ISSN 2410-9754
hsa-miR-24
hsa-miR-548i
hsa-miR-511
hsa-miR-548c-5p
hsa-miR-513a-3p
hsa-miR-548n
hsa-miR-24
hsa-miR-449a
hsa-miR-548i
hsa-miR-511
hsa-miR-545*
hsa-miR-548h
hsa-miR-548b-5p
hsa-miR-548j
hsa-miR-27b
hsa-miR-548i
hsa-miR-27a
hsa-miR-511
hsa-miR-34a
hsa-miR-548h
hsa-miR-338-5p
hsa-miR-548i
hsa-miR-548h
hsa-miR-548d-5p
hsa-miR-454
hsa-miR-548a-5p
hsa-miR-513a-3p
hsa-miR-548h
hsa-miR-548a-5p
hsa-miR-513a-3p
hsa-miR-1243
hsa-miR-576-5p
hsa-miR-548h
hsa-miR-511
hsa-miR-513a-5p
hsa-miR-548d-5p
hsa-miR-891b
hsa-miR-24
hsa-miR-449b
hsa-miR-548i
hsa-miR-511
hsa-miR-548c-5p
hsa-miR-7
hsa-miR-513a-3p
hsa-miR-889
@2014, GNP
Vol:1, 2014
hsa-mir-24-1
hsa-mir-548i-1
hsa-mir-511-1
hsa-mir-548c
hsa-mir-513a-2
hsa-mir-548n
hsa-mir-24-2
hsa-mir-449a
hsa-mir-548i-2
hsa-mir-511-2
hsa-mir-545
hsa-mir-548h-1
hsa-mir-548b
hsa-mir-548j
hsa-mir-27b
hsa-mir-548i-3
hsa-mir-27a
hsa-mir-511-2
hsa-mir-34a
hsa-mir-548h-2
hsa-mir-338
hsa-mir-548i-4
hsa-mir-548h-3
hsa-mir-548d-1
hsa-mir-454
hsa-mir-548a-3
hsa-mir-513a-1
hsa-mir-548h-4
hsa-mir-548a-3
hsa-mir-513a-1
hsa-mir-1243
hsa-mir-576
hsa-mir-548h-4
hsa-mir-511-1
hsa-mir-513a-2
hsa-mir-548d-2
hsa-mir-891b
hsa-mir-24-1
hsa-mir-449b
hsa-mir-548i-1
hsa-mir-511-1
hsa-mir-548c
hsa-mir-7-1
hsa-mir-513a-2
hsa-mir-889
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
8
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
1725
1880
1863
1880
1790
1880
1725
1731
1880
1863
1793
1880
1880
1880
1797
1880
1797
1863
1731
1880
1852
1880
1880
1880
1757
1880
1790
1880
1879
1789
1751
1828
1879
1862
1797
1879
1754
1724
1730
1879
1862
1879
1748
1789
1888
1
1
1
1
1
2
1
1
1
1
2
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
2
2
1
1
2
2
1
2
1
2
2
2
2
2
1
2
1
Biojournal of Science and Technology
1718
1873
1856
1873
1783
1873
1718
1724
1873
1856
1786
1873
1873
1873
1790
1873
1790
1856
1724
1873
1845
1873
1873
1873
1750
1873
1783
1873
1873
1783
1745
1822
1873
1856
1791
1873
1748
1718
1724
1873
1856
1873
1742
1783
1882
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0032
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
Pa g e |5
ISSN 2410-9754
hsa-miR-586
hsa-mir-586
hsa-miR-24
hsa-mir-24-2
hsa-miR-128
hsa-mir-128-2
hsa-miR-7
hsa-mir-7-2
hsa-miR-340
hsa-mir-340
hsa-miR-449a
hsa-mir-449a
hsa-miR-548i
hsa-mir-548i-2
hsa-miR-511
hsa-mir-511-2
hsa-miR-7
hsa-mir-7-3
hsa-miR-548h
hsa-mir-548h-1
hsa-miR-656
hsa-mir-656
hsa-miR-301b
hsa-mir-301b
hsa-miR-548b-5p
hsa-mir-548b
hsa-miR-548j
hsa-mir-548j
hsa-miR-34c-5p
hsa-mir-34c
hsa-miR-27b
hsa-mir-27b
hsa-miR-548i
hsa-mir-548i-3
hsa-miR-27a
hsa-mir-27a
hsa-miR-511
hsa-mir-511-2
hsa-miR-548k
hsa-mir-548k
hsa-miR-34a
hsa-mir-34a
hsa-miR-548h
hsa-mir-548h-2
hsa-miR-128
hsa-mir-128-1
hsa-miR-590-3p
hsa-mir-590
hsa-miR-301a
hsa-mir-301a
hsa-miR-338-5p
hsa-mir-338
hsa-miR-409-3p
hsa-mir-409
hsa-miR-548i
hsa-mir-548i-4
hsa-miR-513a-5p
hsa-mir-513a-1
hsa-miR-130b
hsa-mir-130b
hsa-miR-335*
hsa-mir-335
hsa-miR-548h
hsa-mir-548h-3
hsa-miR-130a
hsa-mir-130a
hsa-miR-1279
hsa-mir-1279
hsa-miR-548l
hsa-mir-548l
hsa-miR-548d-5p
hsa-mir-548d-1
hsa-miR-454
hsa-mir-454
miRNA: microRNA; hsa: Homo sapiens
Vol:1, 2014
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
7
1847
1724
1796
1748
1857
1730
1879
1862
1748
1879
1886
1756
1879
1879
1730
1796
1879
1796
1862
1880
1730
1879
1796
1894
1756
1851
1736
1879
1797
1756
1800
1879
1756
1832
1880
1879
1756
1
2
1
1
1
2
2
2
1
2
1
2
2
2
2
2
2
2
2
1
2
2
1
1
2
2
2
2
1
2
1
2
2
1
1
2
2
1841
1718
1790
1742
1851
1724
1873
1856
1742
1873
1880
1750
1873
1873
1724
1790
1873
1790
1856
1874
1724
1873
1790
1888
1750
1845
1730
1873
1791
1750
1794
1873
1750
1826
1874
1873
1750
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
0.0128
Using miRWalk, number of potential target sites
region) of PPARG in the human genome. The
for miRNAs were identified within the sequences
functional regions of the PPARG gene cluster as
of 5′-UTR (5′-untranslated region), CDS (coding
possible sites for miRNA targeting were further
DNA sequence) and 3′ UTR (3′- untranslated
analyzed. A unique target pattern was pointed
@2014, GNP
Biojournal of Science and Technology
Pa g e |6
ISSN 2410-9754
Vol:1, 2014
within the genomic sequences representing the 5′
regulation
UTR, CDS and 3′ UTR of PPARG gene. Specific
potentials.
and
their
expected
therapeutic
sequences within 5′ UTR, CDS and 3′ UTR of
human PPARG gene along with seed sequences, its
REFERENCES
location and size respectively are shown in tables 1,
1. Bartel DP. 2004. MicroRNAs: genomics,
2 and 3. These experimental data show that the
biogenesis, mechanism, and function. Cell 116:
number of miRNA target sites ranges differently in
281-297.
different regions of PPARG. In the 5′ UTR of the
2. Dweep H, Sticht C, Gretz N. 2013. In-Silico
screened gene, we found 29 different miRNA
Algorithms for the Screening of Possible
target sites with different p values. Among them,
microRNA
the target site for miRNA-181a-2 had the lowest p
Interactions. Curr Genomics 14: 127-136.
value (0.003), i.e. most significant value (Table 1).
3. Dweep H, Sticht C, Pandey P, Gretz N. 2011.
In case of CDS, we obtained 52 target sites,
miRWalk--database: prediction of possible
miRNA-367 being the most significant one (p
miRNA binding sites by "walking" the genes
value= 0.0014) (Table 2). Finally, 85 different
of three genomes. J Biomed Inform 44:
miRNA target sites were identified within the 3′
839-847.
Binding
Sites
and
Their
UTR. We found miRNA-559 be the most
4. Friedman RC, Farh KK, Burge CB, Bartel DP.
significant one (p= 0.0080 amongst all within this
2009. Most mammalian mRNAs are conserved
region (Table 3). The findings would help when we
targets of microRNAs. Genome Res 19:
want to select miRNAs for studying their role in
92-105.
PPARG regulation in laboratory conditions.
5. He L, Hannon GJ. 2004. MicroRNAs: small
RNAs with a big role in gene regulation. Nat
A
number
of
computational
miRNA-target
prediction algorithms have been developed due to
Rev Genet 5: 522-531.
6. John B, Enright AJ, Aravin A, Tuschl T,
lack of high-throughput experimental methods but
Sander
C,
Marks
DS.
2004.
Human
these programs still lacking sensitivity and
MicroRNA targets. PLoS Biol 2: e363.
specificity. The miRWalk database provides a
7. Kiriakidou M, Nelson PT, Kouranov A, Fitziev
comprehensive atlas of putative miRNA binding
P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou
site prediction from multiple algorithms and
A.
therefore attracts researchers. These existing
computational-experimental approach predicts
algorithms will become more accurate with more
human microRNA targets. Genes Dev 18:
understanding of miRNA regulatory mechanism
1165-1178.
(Dweep et al. 2013). It can thus be concluded that
a
combination
both
computational
A
combined
8. Landgraf P, et al. 2007. A mammalian
and
microRNA expression atlas based on small
experimental approaches would be required to
RNA library sequencing. Cell 129: 1401-1414.
unravel the complex networks of miRNA gene
9. Lewis BP, Shih IH, Jones-Rhoades MW,
@2014, GNP
of
2004.
Biojournal of Science and Technology
Pa g e |7
ISSN 2410-9754
Vol:1, 2014
Bartel DP, Burge CB. 2003. Prediction of
17. Vidal-Puig A, Jimenez-Linan M, Lowell BB,
mammalian microRNA targets. Cell 115:
Hamann A, Hu E, Spiegelman B, Flier JS,
787-798.
Moller DE. 1996. Regulation of PPAR gamma
10. Li Y, Qi Y, Huang TH, Yamahara J, Roufogalis
BD. 2008. Pomegranate flower: a unique
traditional antidiabetic medicine with dual
PPAR-alpha/-gamma
activator
properties.
Diabetes Obes Metab 10: 10-17.
gene expression by nutrition and obesity in
rodents. J Clin Invest 97: 2553-2561.
18. Vidal-Puig AJ, Considine RV, Jimenez-Linan
M, Werman A, Pories WJ, Caro JF, Flier JS.
1997.
Peroxisome
proliferator-activated
11. Martin G, Schouest K, Kovvuru P, Spillane C.
receptor gene expression in human tissues.
2007. Prediction and validation of microRNA
Effects of obesity, weight loss, and regulation
targets in animal genomes. J Biosci 32:
by insulin and glucocorticoids. J Clin Invest
1049-1052.
99: 2416-2422.
12. Perron
MP,
Provost
interactions
and
P.
2008.
complexes
in
Protein
19. Zeng Y, Cullen BR. 2006. Recognition and
human
cleavage of primary microRNA transcripts.
microRNA biogenesis and function. Front
Methods Mol Biol 342: 49-56.
Biosci 13: 2537-2547.
13. Rajewsky N, Socci ND. 2004. Computational
identification of microRNA targets. Dev Biol
267: 529-535.
14. Rosen ED, Sarraf P, Troy AE, Bradwin G,
Moore K, Milstone DS, Spiegelman BM,
Mortensen RM. 1999. PPAR gamma is
required for the differentiation of adipose
tissue in vivo and in vitro. Mol Cell 4:
611-617.
15. Schoonjans K, Peinado-Onsurbe J, Lefebvre
AM, Heyman RA, Briggs M, Deeb S, Staels B,
Auwerx J. 1996. PPARalpha and PPARgamma
activators direct a distinct tissue-specific
transcriptional response via a PPRE in the
lipoprotein
lipase
gene.
EMBO
J
15:
5336-5348.
16. Srinivasan S, Selvan ST, Archunan G, Gulyas
B, Padmanabhan P. 2013. MicroRNAs -the
next generation therapeutic targets in human
diseases. Theranostics 3: 930-942.
@2014, GNP
Biojournal of Science and Technology
Pa g e |8
Download m140002
m140002.pdf (PDF, 270.04 KB)
Download PDF
Share this file on social networks
Link to this page
Permanent link
Use the permanent link to the download page to share your document on Facebook, Twitter, LinkedIn, or directly with a contact by e-Mail, Messenger, Whatsapp, Line..
Short link
Use the short link to share your document on Twitter or by text message (SMS)
HTML Code
Copy the following HTML code to share your document on a Website or Blog
QR Code to this page

This file has been shared publicly by a user of PDF Archive.
Document ID: 0000291803.